Members Area
Cavalier King Charles Spaniel
* Available as custom analyses. Contact us for details.
Genetic Diversity
Cavalier genetic diversity
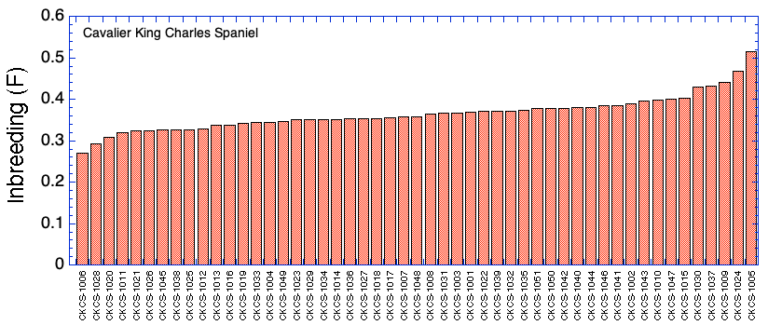
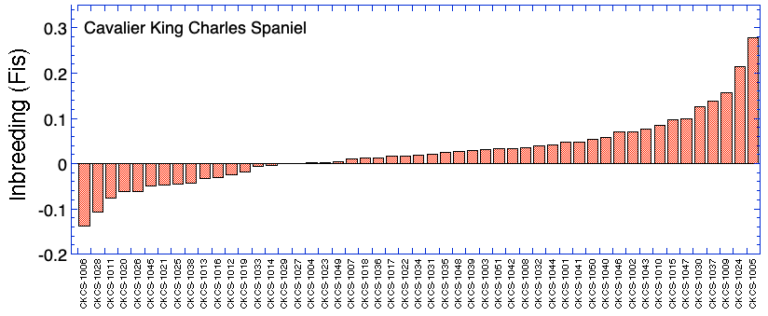
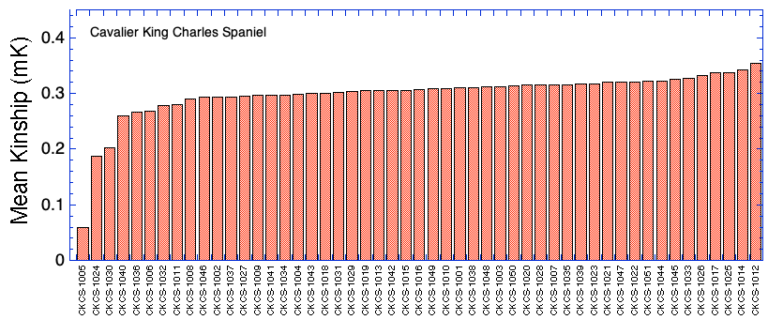
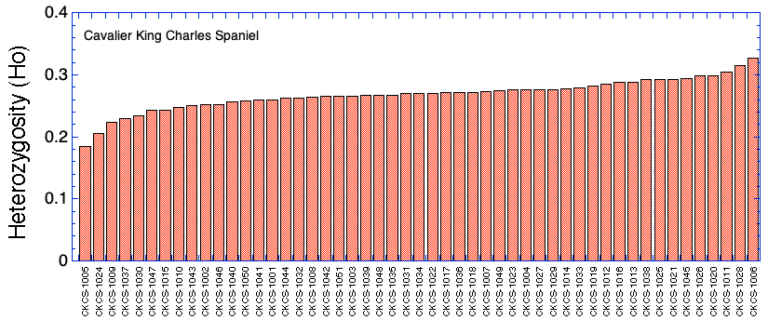
| ID code | Inbreeding (F) | Inbreeding (Fis) | Mean Kinship (mK) | Heterozygosity (Ho) | |
|---|---|---|---|---|---|
| wdt_ID | ID code | Inbreeding (F) | Inbreeding (Fis) | Mean Kinship (mK) | Heterozygosity (Ho) |
| 1 | CKCS-1001 | 0.368 | 0.048 | 0.310 | 0.260 |
| 2 | CKCS-1002 | 0.390 | 0.070 | 0.293 | 0.252 |
| 3 | CKCS-1003 | 0.367 | 0.032 | 0.312 | 0.265 |
| 4 | CKCS-1004 | 0.345 | 0.002 | 0.299 | 0.276 |
| 5 | CKCS-1005 | 0.515 | 0.279 | 0.059 | 0.185 |
| 6 | CKCS-1006 | 0.269 | -0.138 | 0.268 | 0.327 |
| 7 | CKCS-1007 | 0.357 | 0.011 | 0.316 | 0.272 |
| 8 | CKCS-1008 | 0.366 | 0.036 | 0.291 | 0.264 |
| 9 | CKCS-1009 | 0.441 | 0.157 | 0.297 | 0.223 |
| 10 | CKCS-1010 | 0.399 | 0.084 | 0.309 | 0.247 |
| 11 | CKCS-1011 | 0.319 | -0.076 | 0.280 | 0.304 |
| 12 | CKCS-1012 | 0.328 | -0.024 | 0.355 | 0.285 |
| 13 | CKCS-1013 | 0.337 | -0.033 | 0.306 | 0.288 |
| 14 | CKCS-1014 | 0.351 | -0.005 | 0.342 | 0.278 |
| 15 | CKCS-1015 | 0.404 | 0.097 | 0.306 | 0.243 |
| 16 | CKCS-1016 | 0.337 | -0.032 | 0.307 | 0.288 |
| 17 | CKCS-1017 | 0.357 | 0.016 | 0.337 | 0.271 |
| 18 | CKCS-1018 | 0.354 | 0.013 | 0.301 | 0.272 |
| 19 | CKCS-1019 | 0.342 | -0.018 | 0.305 | 0.283 |
| 20 | CKCS-1020 | 0.309 | -0.062 | 0.316 | 0.299 |
| 21 | CKCS-1021 | 0.323 | -0.046 | 0.320 | 0.293 |
| 22 | CKCS-1022 | 0.371 | 0.016 | 0.321 | 0.271 |
| 23 | CKCS-1023 | 0.350 | 0.003 | 0.317 | 0.275 |
| 24 | CKCS-1024 | 0.468 | 0.215 | 0.187 | 0.205 |
| 25 | CKCS-1025 | 0.327 | -0.045 | 0.338 | 0.292 |
| 26 | CKCS-1026 | 0.324 | -0.061 | 0.333 | 0.298 |
| 27 | CKCS-1027 | 0.353 | 0.000 | 0.295 | 0.276 |
| 28 | CKCS-1028 | 0.292 | -0.108 | 0.316 | 0.316 |
| 29 | CKCS-1029 | 0.350 | 0.000 | 0.304 | 0.276 |
| 30 | CKCS-1030 | 0.429 | 0.126 | 0.202 | 0.234 |
| 31 | CKCS-1031 | 0.366 | 0.021 | 0.302 | 0.269 |
| 32 | CKCS-1032 | 0.372 | 0.040 | 0.279 | 0.263 |
| 33 | CKCS-1033 | 0.345 | -0.006 | 0.328 | 0.279 |
| 34 | CKCS-1034 | 0.350 | 0.020 | 0.297 | 0.270 |
| 35 | CKCS-1035 | 0.374 | 0.026 | 0.316 | 0.267 |
| 36 | CKCS-1036 | 0.353 | 0.013 | 0.266 | 0.272 |
| 37 | CKCS-1037 | 0.433 | 0.137 | 0.294 | 0.230 |
| 38 | CKCS-1038 | 0.327 | -0.043 | 0.310 | 0.292 |
| 39 | CKCS-1039 | 0.372 | 0.029 | 0.317 | 0.266 |
| 40 | CKCS-1040 | 0.380 | 0.058 | 0.259 | 0.256 |
| 41 | CKCS-1041 | 0.385 | 0.048 | 0.297 | 0.260 |
| 42 | CKCS-1042 | 0.379 | 0.033 | 0.306 | 0.265 |
| 43 | CKCS-1043 | 0.396 | 0.077 | 0.301 | 0.250 |
| 44 | CKCS-1044 | 0.381 | 0.041 | 0.323 | 0.262 |
| 45 | CKCS-1045 | 0.327 | -0.050 | 0.326 | 0.294 |
| 46 | CKCS-1046 | 0.385 | 0.070 | 0.293 | 0.252 |
| 47 | CKCS-1047 | 0.401 | 0.098 | 0.320 | 0.243 |
| 48 | CKCS-1048 | 0.357 | 0.028 | 0.312 | 0.267 |
| 49 | CKCS-1049 | 0.347 | 0.004 | 0.308 | 0.275 |
| 50 | CKCS-1050 | 0.379 | 0.053 | 0.313 | 0.258 |
| 51 | CKCS-1051 | 0.377 | 0.033 | 0.322 | 0.265 |
Traits & Disorders
(no data available)
Sorted by rank
Ranking by Inbreeding (F)
Ranking by Inbreeding (Fis)
Ranking by Mean Kinship
Ranking by Heterozygosity
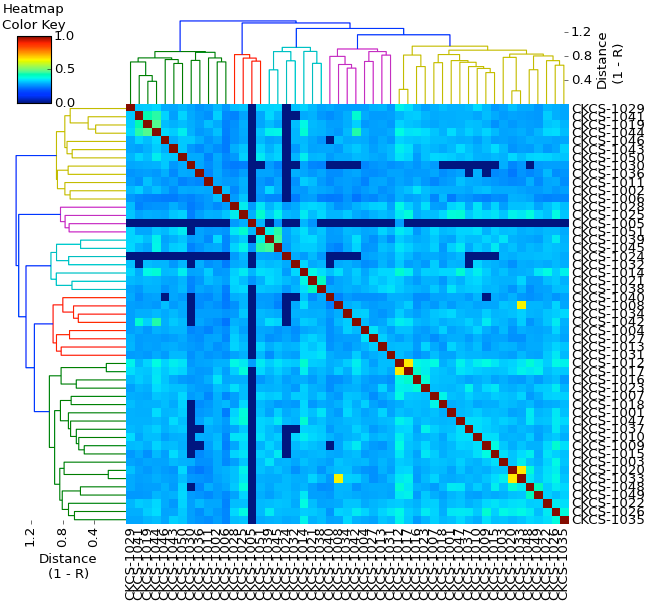
Genetic Structure
The dendrogram and heat map for genetic structure are produced from the kinship coefficients.
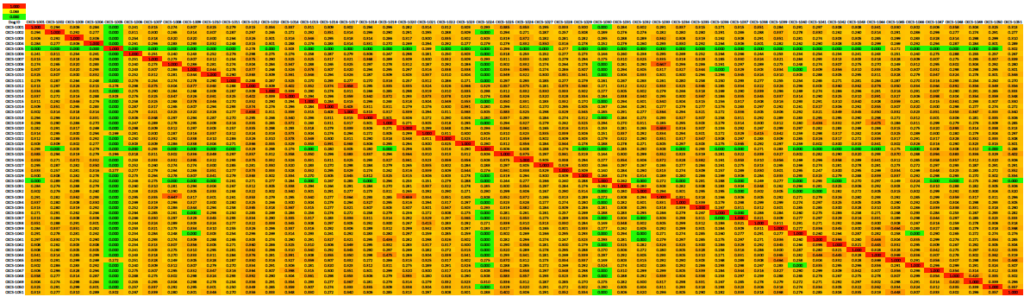
Kinship Coefficient
The kinship coefficient (K) is the fraction of the genome shared by two individuals that is homozygous by descent, so it is a pairwise measure of relatedness.
This kinship matrix is color coded to identify cells in which K < 0.0625 (6.25%; green, equivalent to a cross of first cousins), K = 0.125 (12.5%; yellow, half-sib cross), and K > 0.25 (25%; red, full sib cross). The kinship coefficient between a male and female is also the predicted average inbreeding coefficient of their offspring if they produced a litter.
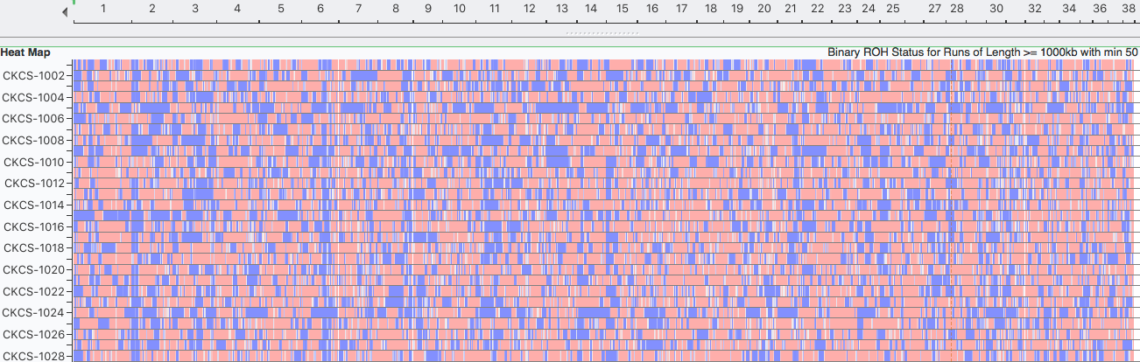
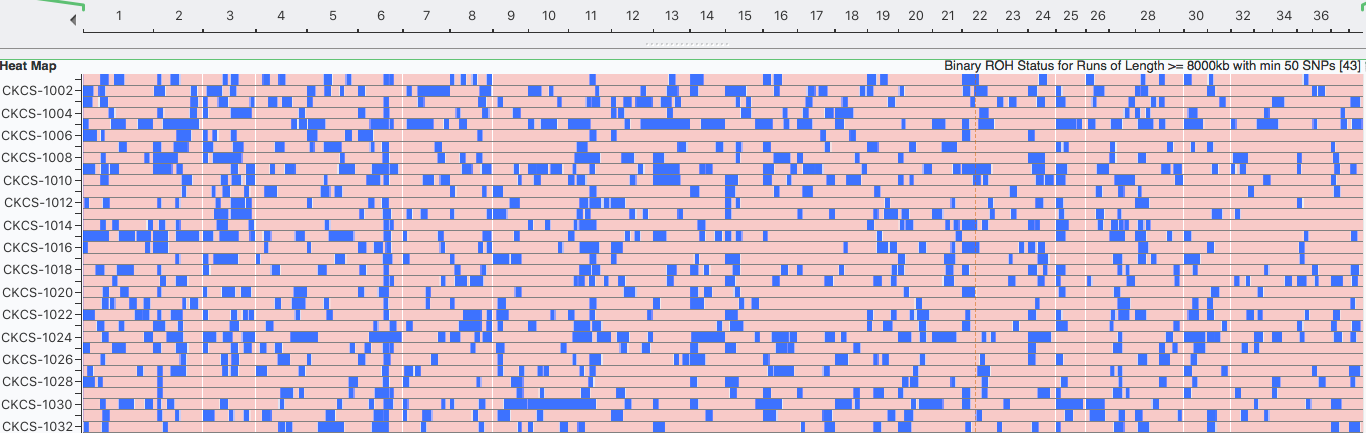
Runs of Homozygosity (ROH)
Inbreeding tends to produce runs (blocks) of homozygosity (ROH) on the chromosomes. Over time, the crossing-over of chromosomes during meiosis can split up blocks of homozygosity, so the largest blocks are likely to be the most recent inbreeding and smaller blocks reflect older inbreeding. ROH are used to estimate inbreeding from SNP data.
Learn more about Runs of Homozygosity.
Inbreeding: 25 generations (1000 kb)
Inbreeding: 6 generations (8000 kb)
(no data available)
(no data available)